Release Party
Mighty Metrika Interface to Cluster Adjusted t Statistics
The goal of the 'mmiCATs' R package is to introduce researchers to cluster adjusted t statistics (CATs), help them better understand when to use them, and to provide a tool which makes the method easy to get started using.
My Introduction to CATs
As you may already know, mightymetrika's mission is centered around advancing methods for statistical methods which improve the analysis of small sample size data. A lot of studies that I work on have a double complication:
- Small number of participants
- A clustering structure such as repeated measurements within participant
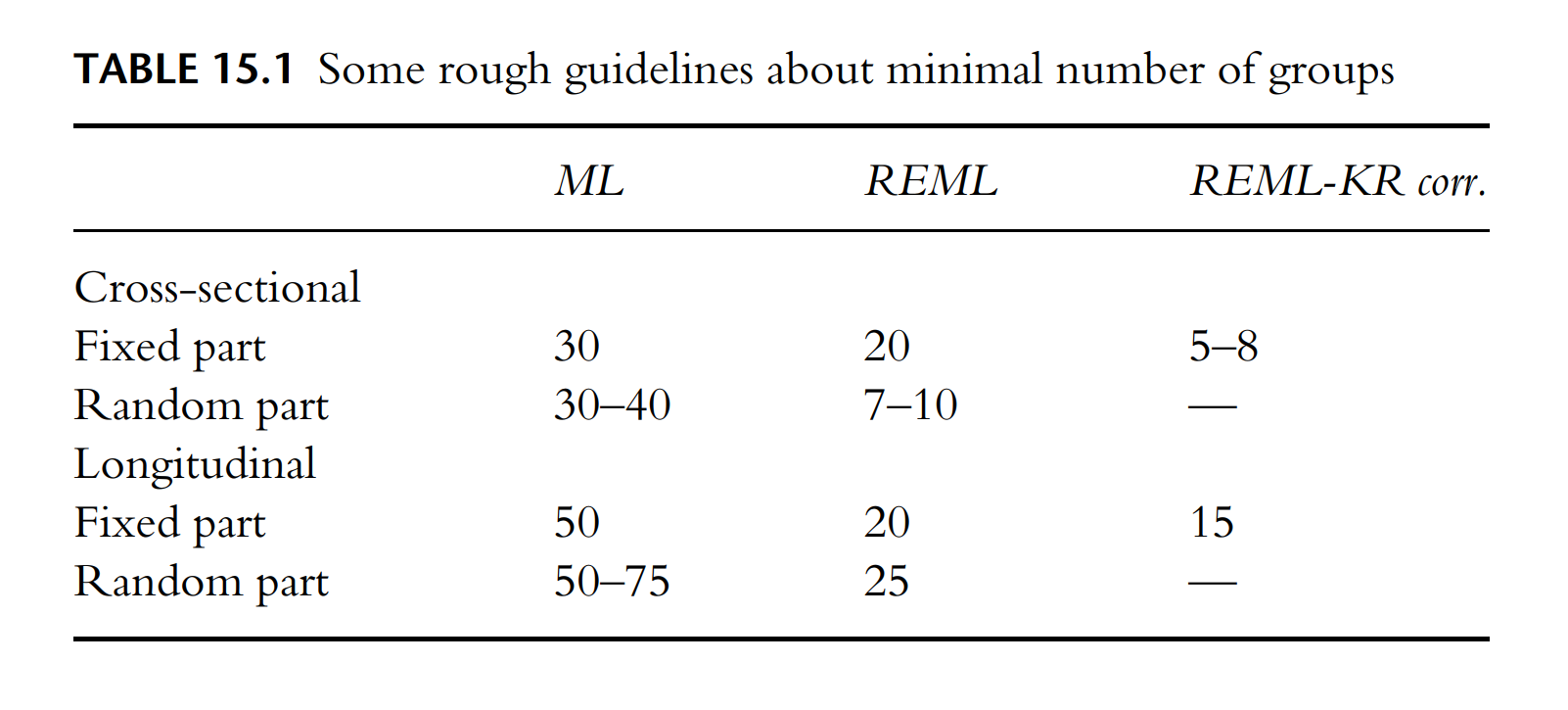
When looking for papers that discuss methods for handling this double complication, I came across Small Samples in Multilevel Modeling by Joop Hox & Daniel McNeish (2020). I often reference this paper in my work as an applied statistician. In particular, I often find two key parts of the paper extremely helpful. The first part that I often return to is Table 15.1 (see image below) and the discussion surrounding.
The other part of the paper which I find very useful is the discussion of what results from Bayesian models might look like in comparison to the results presented in Table 15.1:
"Table 15.1 does not mention Bayesian estimation because suggestions are highly dependent on specification of the prior. With uninformative priors, Bayesian estimation should work with the sample sizes indicated for ML. Bayesian estimation with weakly informative priors roughly corresponds to the REML column, and Bayesian estimation with strongly informative priors is typically appropriate with lower samples than suggested for REML with the Kenward–Roger correction (McNeish, 2016b). "
Originally, I read this paper as an applied statistician looking for more efficient ways to implement mixed effects modeling with smaller sample sizes. However, when I started working on mightymetrika, I came to a point where I wanted to work with a method for smaller sample sizes when the data generating process suggests that the "independent observations" assumption of a statistical model is violated; however, I was not ready to start working on a project focused on prior distributions. So I came back and read the Hox & McNeish (2020) paper again, and this time, here is the passage that stood out for me:
A study by Cameron, Gelbach, and Miller (2008) showed that the “wild bootstrap”, which is similar to the residual bootstrap, was effective with as few as five clusters, which is even lower that the minimal sample size reported in Yung and Chan (1999)...the wild bootstrap can be carried out in the R package 'clusterSEs' (Esarey & Menger, 2018).
To learn more about this fascinating wild bootstrap method, I turned to the Esarey & Menger (2018) paper which studies four methods side-by-side:
- Cluster-robust standard error
- Which might be considered the reference method which inspires the search for a replacement
- Pairs cluster bootstrapped t-statistics
- Wild cluster bootstrapped t-statistics
- Which I came to learn about
- Cluster-adjusted t-statistics
- Which turned out to be the star
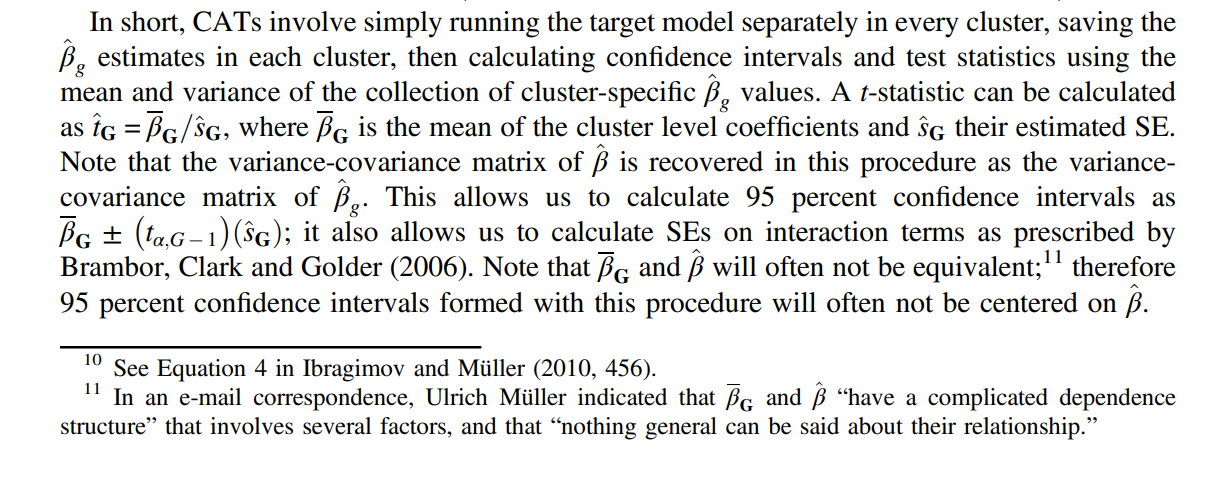
One section of the paper provides a relatively simple overview of the CATs method:
When to Use CATs
The conclusion section of Esarey & Menger (2018) had a paragraph which provided a nice outline of when to select which method:
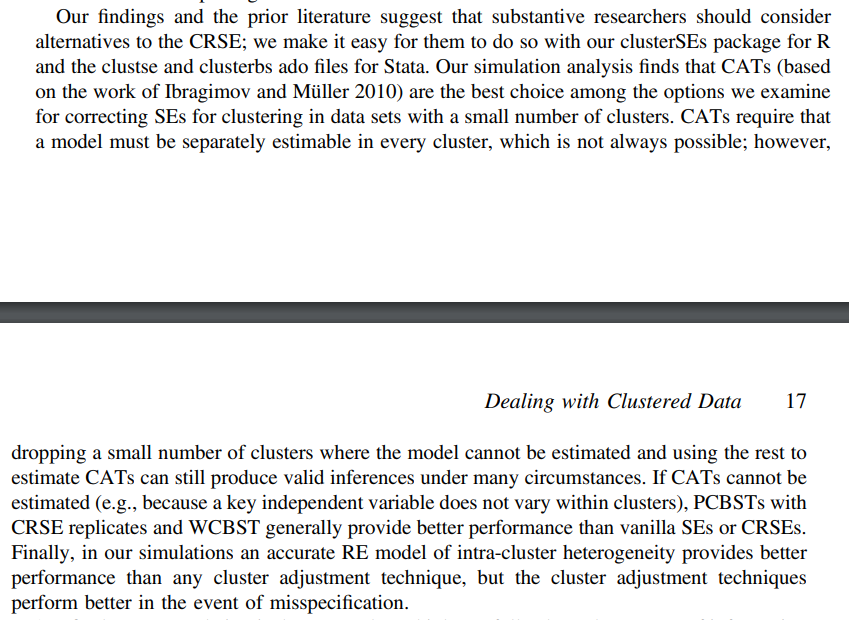
Often, researchers select a random effects model to handle clustered data. However, when the sample size is small, a statistician might try to avoid specifying a model that is too complex for the data by specifying a simpler random effects model than the model considered to be the correct specification. The 'mmiCATs' R package comes with a card game called CloseCATs(). The 'mmiCATs' GitHub README describes the game as follows:

How to Start Using CATs
To start using CATs, I recommend three resources:
- Esarey & Menger (2018) for a better understanding of the method
- The clusterSEs R package which you can find on CRAN
- The MetaCran GitHub repository for clusterSEs where you can browse the code for the method
If you have a csv with clustered data and you would like to get started experimenting with CATs, you can also use the mmiCATs::mmiCATs() shiny application on the mightymetrika website. This app implements the basic functionality of the clusterSEs::clusterIM.glm() function.
Coming Soon
In the coming weeks, mightymetrika.com is planning to release a few blog posts which can help you delve deeper into CATs including:
- A tutorial which will use screenshots to walk through a CATs analysis using the mmiCATs::mmiCATs() application
- A blog post that will walk through a hand or two of the mmiCATs::CloseCATs() game
In addition to the two main 'mmiCATs' functions (mmiCATs() & CloseCATs()), the 'mmiCATs' package also includes two functions which experiment with the possibility of using CATs with robust regression models in place of stats::glm():
- mmiCATs::cluster_im_lmRob()
- mmiCATs::cluster_im_glmRob()
These functions can be used with robust models from both 'robust' and 'robustbase'.
The mmiCATs::pwr_func_lmer() function can be used to streamline the workflow of conducting simulation studies by:
- Simulating clustered datasets
- Fitting the data sets to the following statistical models:
- Linear mixed effects model
- Random intercept model
- CATs
- CATs with truncation
- mmiCATs::cluster_im_lmRob() with 'robust'
- mmiCATs::cluster_im_lmRob() with 'robustbase'
Then results can be compared to give users a better understanding of each models performance under various data generation schemes.


